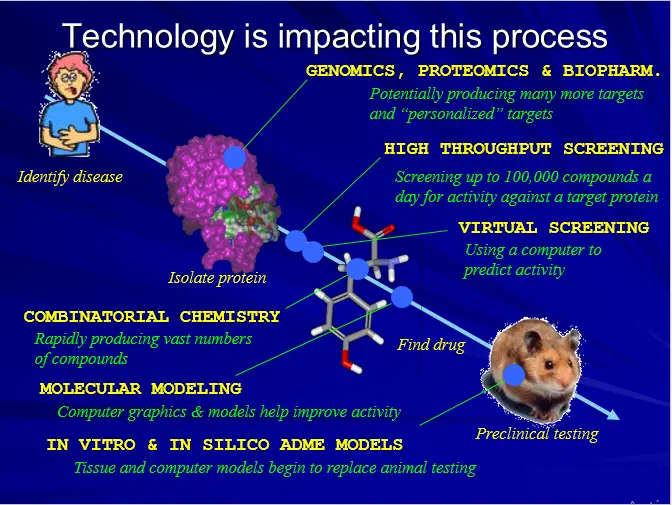
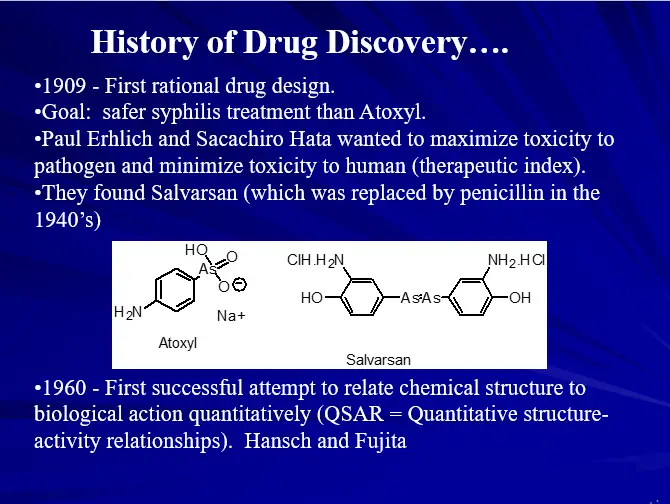
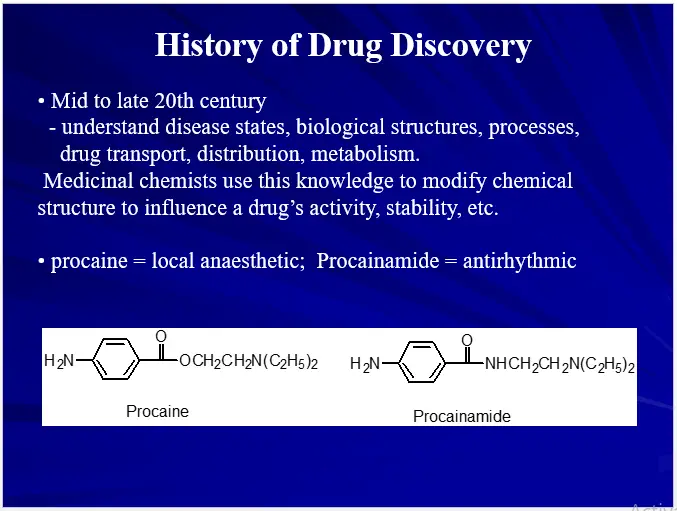
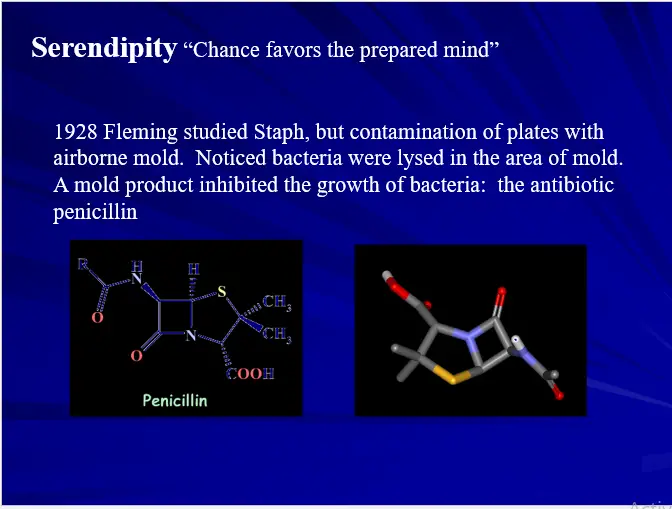
DRUG DESIGN (AN OVERVIEW)
DRUG DESIGN
DRUG DESIGN Approaches to drug discovery:
- Serendipity (luck)
- Chemical Modification
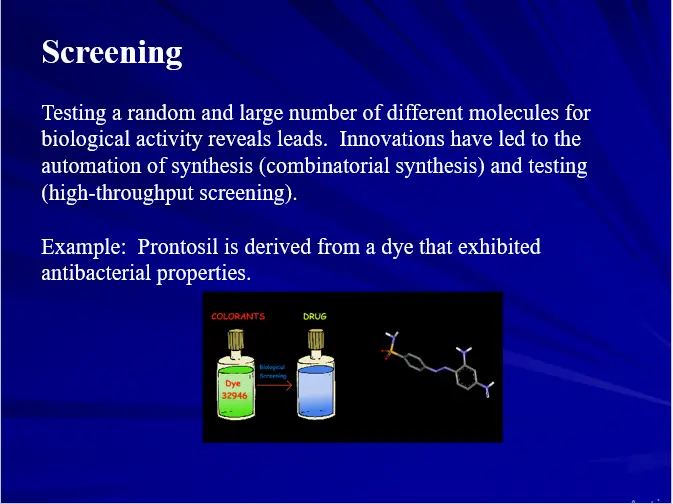
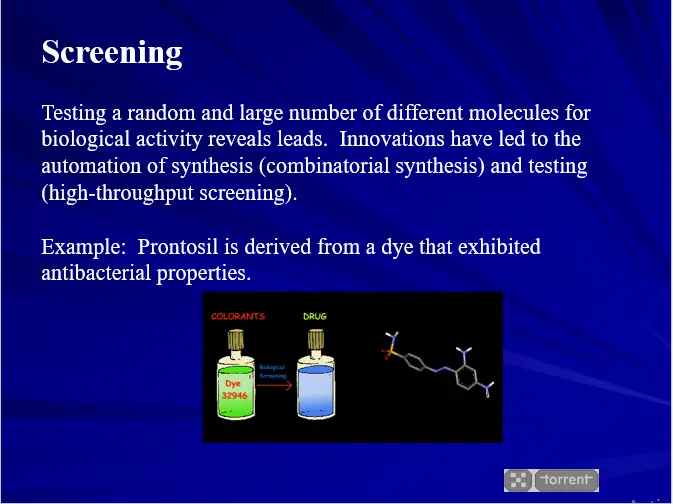
- Screening
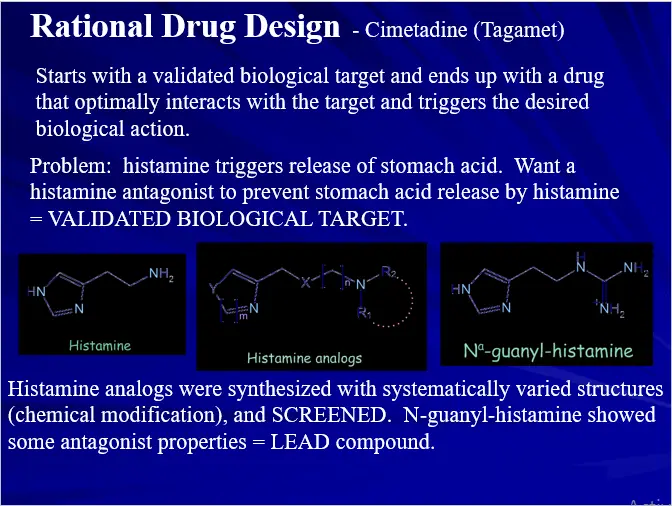
- Rational
Irrational, based on serendipity & Intuition
Trial & error approach in DRUG DESIGN
Time consuming with low through output
No de novo design, mostly “Me Too Approach”
First generation Rational approach in Drug design
In 1970s the medicinal chemists considered molecules as topological entities in 2 dimension (2D) with associated chemical properties.
QSAR concept became quite popular. It was implemented in computers and constituted first generation rational approach to drug design
2nd generation rational drug design
The acceptance by medicinal chemists of molecular modeling was favored by the fact that the QSAR was now supplemented by 3D visualization.
The “lock and key” complementarity is actually supported by 3D model. Computer aided molecular design (CAMD) is expected to contribute to intelligent lead
MECHANISM BASED DRUG-DESIGN
- Most rational approach employed today.
- Disease process is understood at molecular level & targets are well defined.
- Drug can then be designed to effectively bind these targets & disrupt the disease process
- Very complex & intellectual approach & therefore requires detailed knowledge & information retrieval. (CADD Holds Great Future)
“Drug –Receptor Interaction is not merely a lock-key interaction but a dynamic & energetically favorable one”
Evolutionary drug designing
Ancient times: Natural products with biological activities used as drugs.
Chemical Era: Synthetic organic compounds
Rationalizing design process: SAR & Computational Chemistry based Drugs
Biochemical era: To elucidate biochemical pathways and macromolecular structures as target as well as drug.
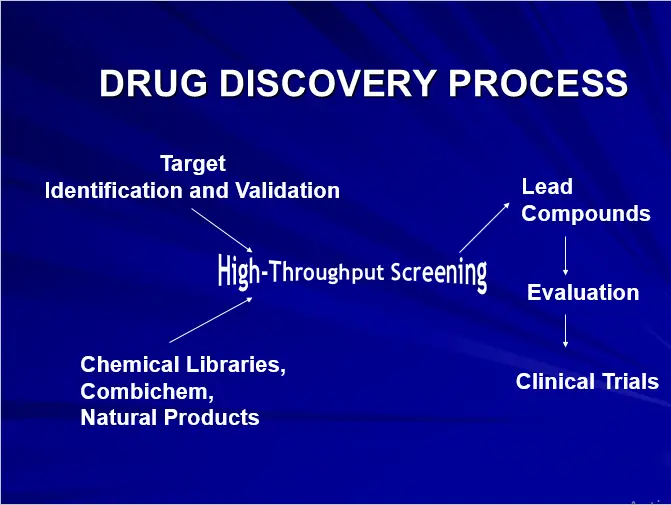
HIGH-THROUGHPUT SCREENING DRUG DESIGN
- FUNCTIONAL INTEGRATION OF:
- BIOLOGY
- CHEMISTRY
- SCREENING TECHNOLOGY
- INFORMATICS
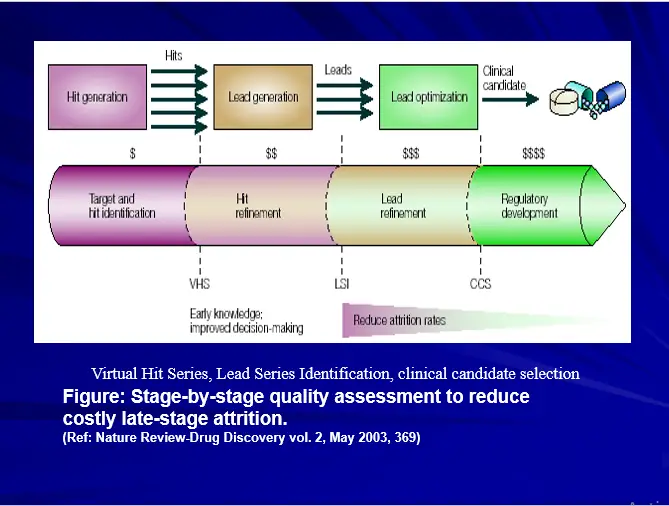
BOTTLENECKS OF DRUG DESIGN
Hundreds of “Hits” but NO “Leads”
Data mining
Accurate profiling of molecules for further studies.
ALTERNATE STRATEGIES OF DRUG DESIGN
Rational Design of Chemical Libraries
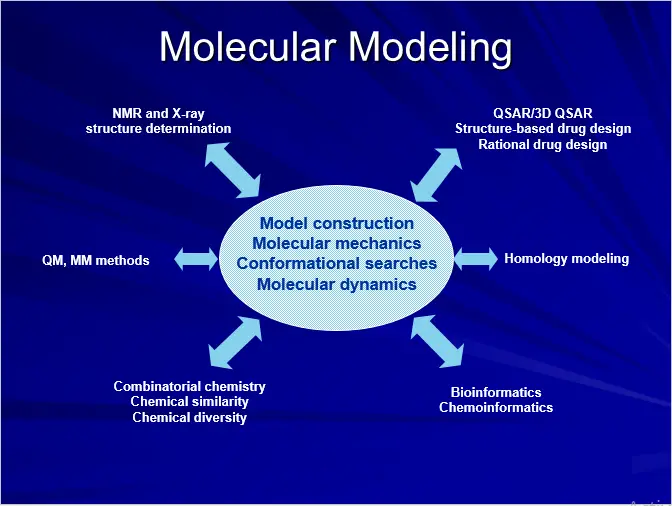
Molecular Modeling Approach
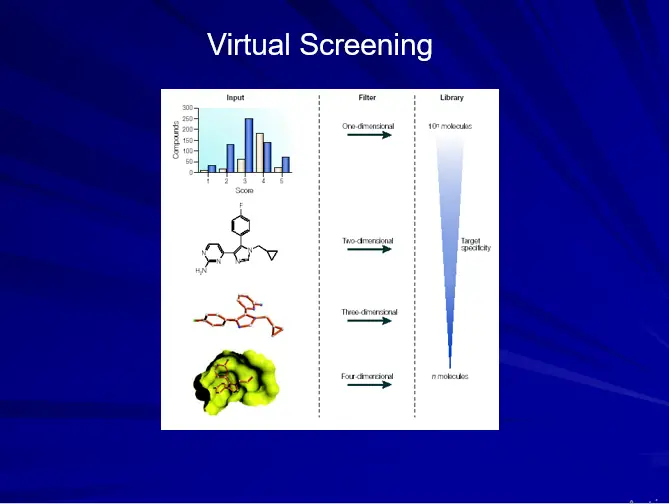
Virtual Screening
Early ADME & Toxicity Profiling
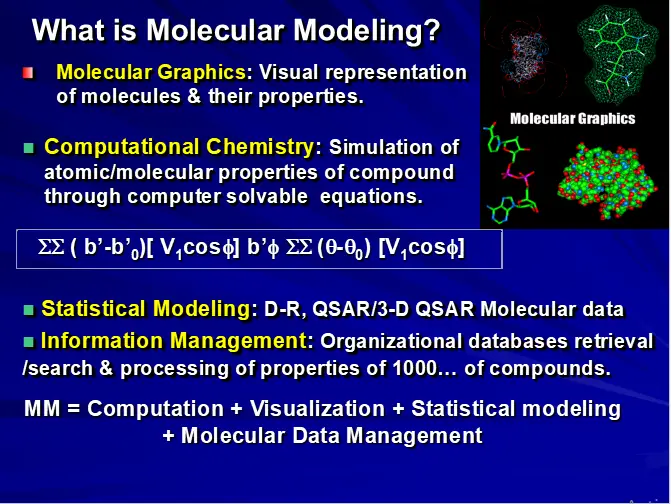
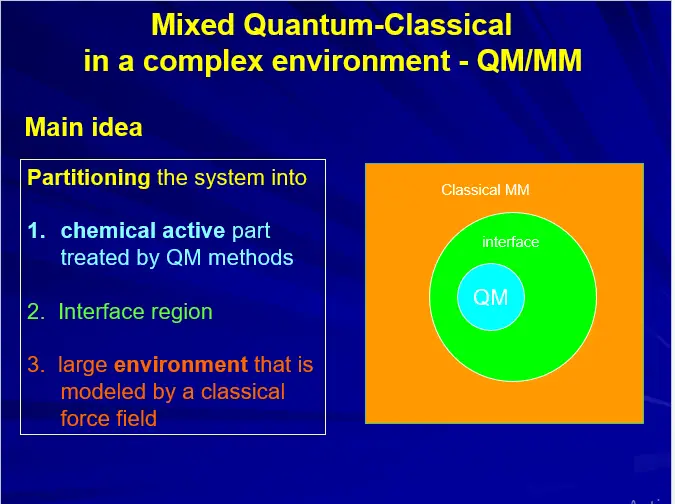
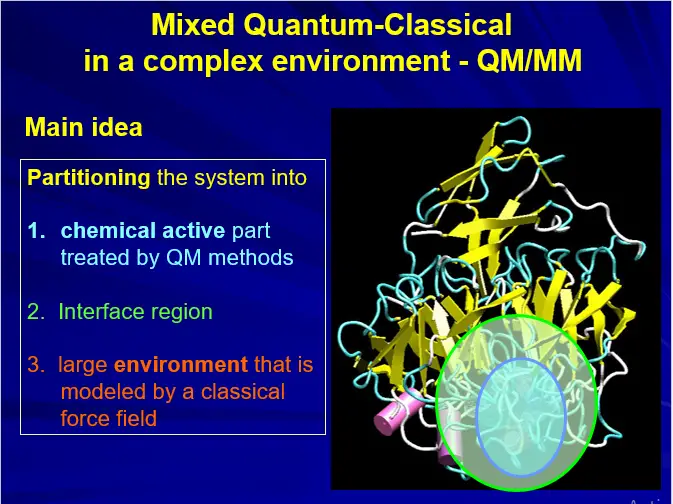
COMPUTATIONAL TOOLS: QM/MM
(A) MOLECULAR MECHANICS (MM)
(B) QUANTUM MECHANICS (QM)
COMPUTATIONAL TOOLS
Quantum Mechanics (QM)
Ab-initio and semi-empirical methods
Considers electronic effect & electronic structure of the molecule
Calculates charge distribution and orbital energies
Can simulate bond breaking and formation
Upper atom limit of about 100-120 atoms
Molecular Mechanics (MM)
Totally empirical technique applicable to both small and macromolecular systems
A molecule is described as a series of charged points (atoms) linked by springs (bonds)
The potential energy of molecule is described by a mathematical function called a FORCE FIELD
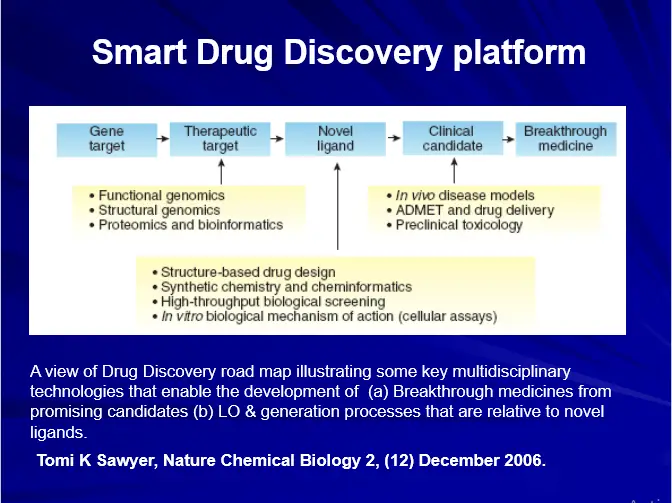
Computer Aided Drug Design Techniques
-Physicochemical Properties Calculations-
– Partition Coefficient (LogP), Dissociation Constant (pKa) etc.
– Drug Design-
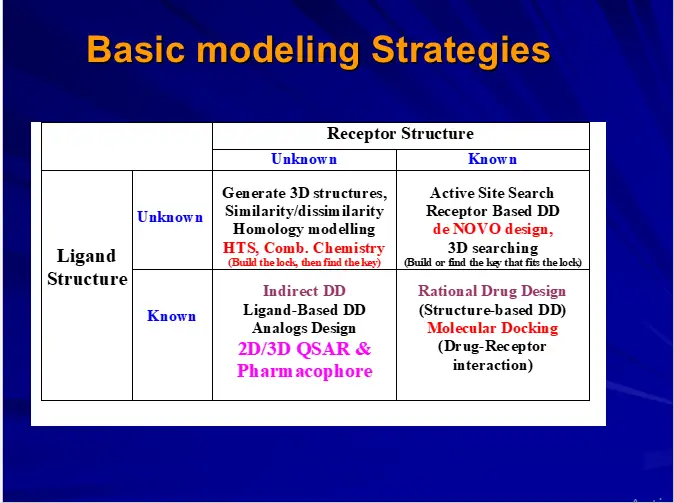
– Ligand Based Drug Design
– QSARs
– Pharmacophore Perception-
– Structure Based Drug Design
– Docking & Scoring
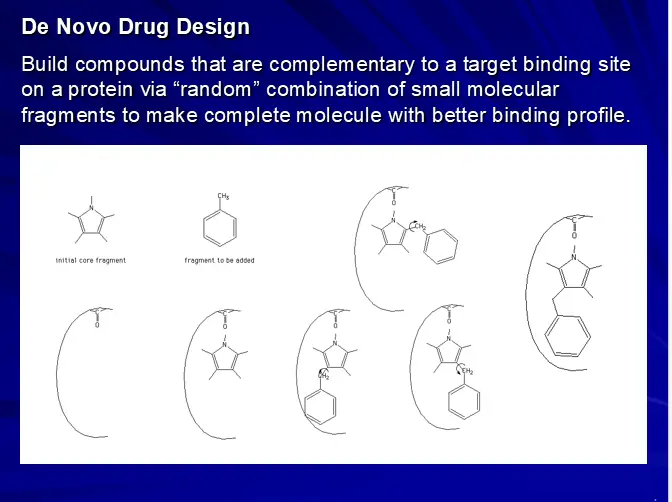
– de-novo drug design
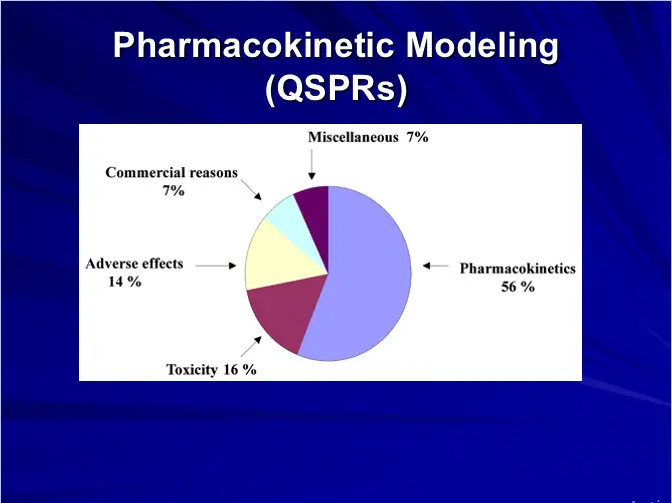
– Pharmacokinetic Modeling (QSPRs)
– Absorption, Metabolism, Distribution and Toxicity etc.
– Cheminformatics
– Database Management
– Similarity / Diversity Searches
-All techniques joins together to form VIRTUAL SCREENING protocols
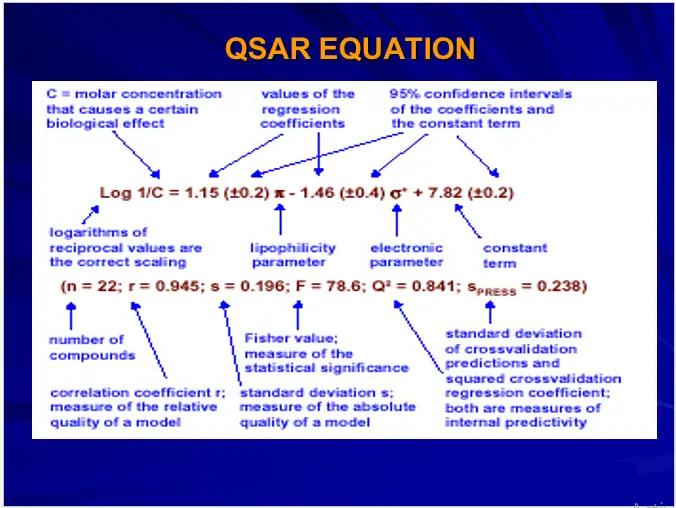
Quantitative Structure Activity Relationships (QSAR)
QSARs are the mathematical relationships linking chemical structures with biological activity using physicochemical or any other derived property as an interface.
Biological Activity = f (Physico-chemical properties)
Mathematical Methods used in QSAR includes various regression and pattern recognition techniques.
Physicochemical or any other property used for generating QSARs is termed as Descriptors and treated as independent variable.
Biological property is treated as dependent variable.
Types of QSARs
Two Dimensional QSAR
– Classical Hansh Analysis
– Two dimensional molecular properties
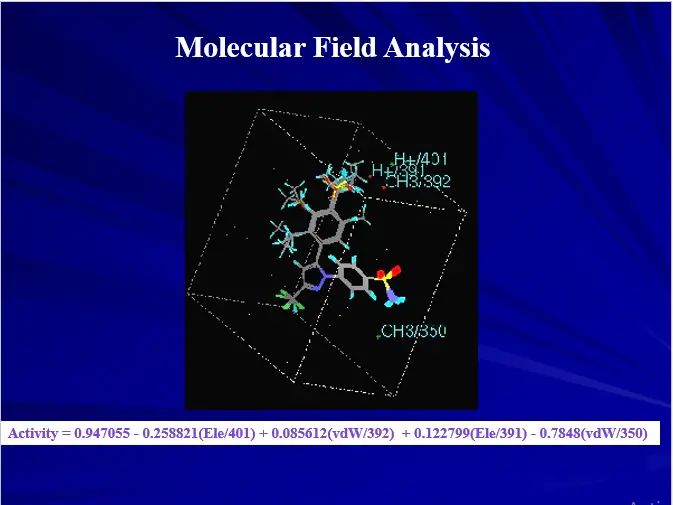
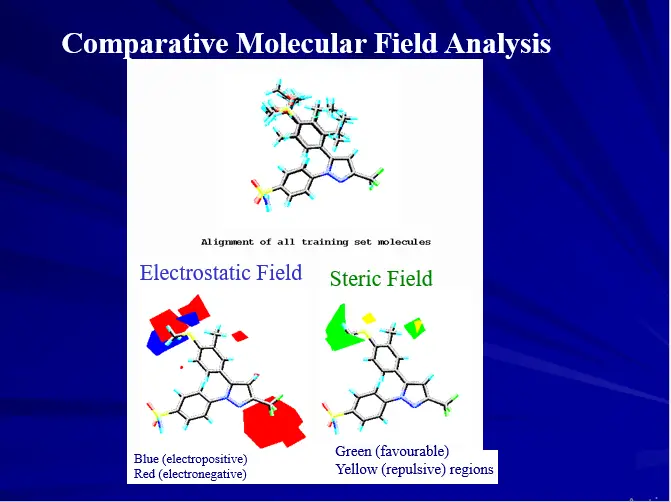
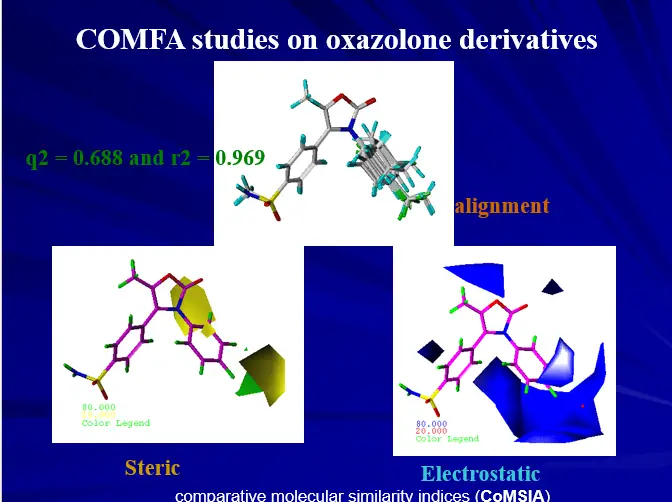
Three Dimensional QSAR
– Three dimensional molecular properties
– Molecular Field Analysis
– Molecular Shape Analysis
– Distance Geometry
– Receptor Surface Analysis
QSAR ASSUMPTIONS
The Effect is produced by model compound and not it’s metabolites.
The proposed conformation is the bioactive one.
The binding site is same for all modeled compounds.
The Bioactivity explain the direct interaction of molecule and target.
Pharmacokinetics aspects, solvent effects, diffusion, transport are not under consideration.
QSAR Generation Process
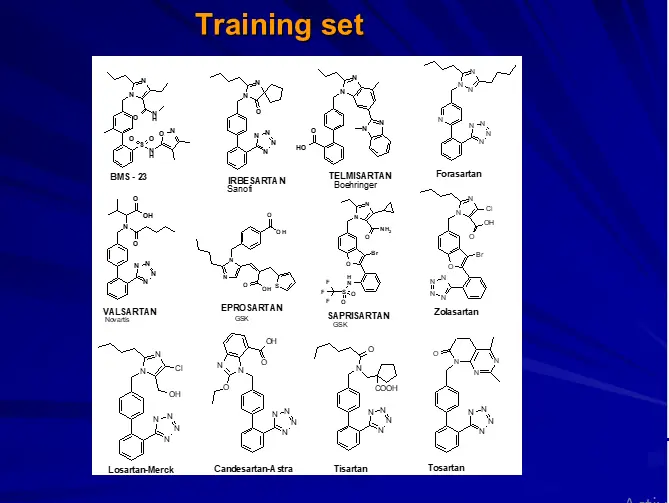
1. Selection of training set
2. Enter biological activity data
3. Generate conformations
4. Calculate descriptors
5. Selection of statistical method
6. Generate a QSAR equation
7. Validation of QSAR equation
8. Predict for Unknown
Descriptors
1.Structural descriptors
2.Electronic descriptors
3.Quantum Mech. descriptors
4.Thermodynamic descriptors
5.Shape descriptors
6.Spatial descriptors
7.Conformational descriptors
8.Receptor descriptors
Selection of Descriptors
1.What is particularly relevant to the therapeutic target?
2.What variation is relevant to the compound series?
3.What property data can be readily measured?
4.What can be readily calculated?
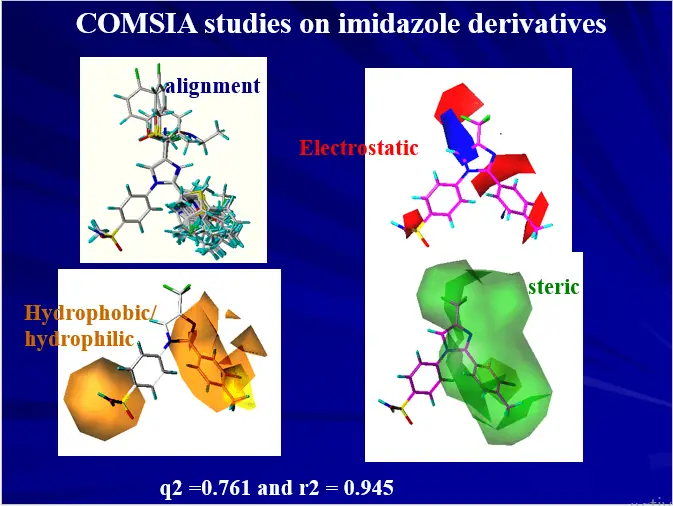
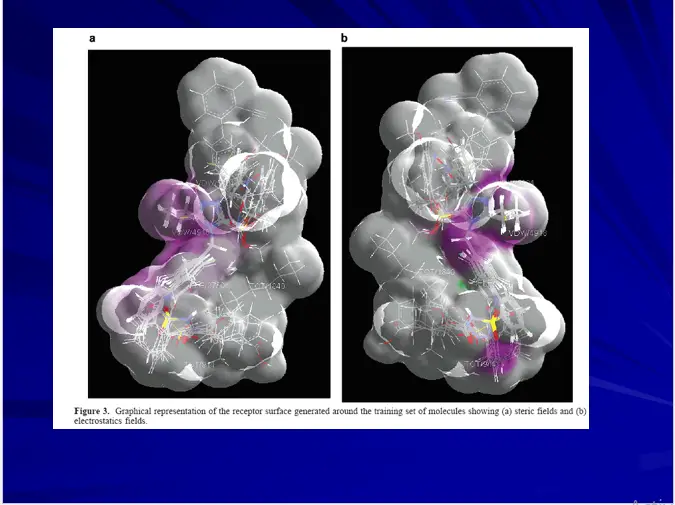
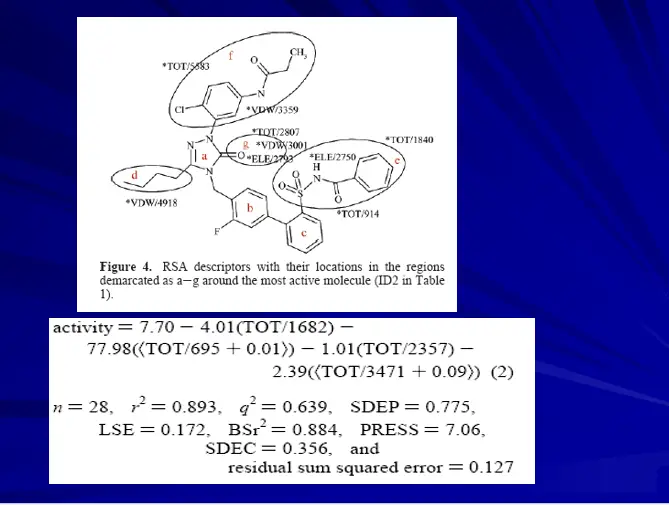
3D-QSAR – RECEPTOR SURFACE MODEL
Hypothetical receptor surface model constructed from training set molecule’s 3D shape and activity data.
The best model can be derived by optimizing various parameters like atomic partial charges and surface fit.
Descriptors like van der Waals energy, electrostatic energy, and total non-bonded energy can be used to derived series of QSAR equations using G/PLS statistical method
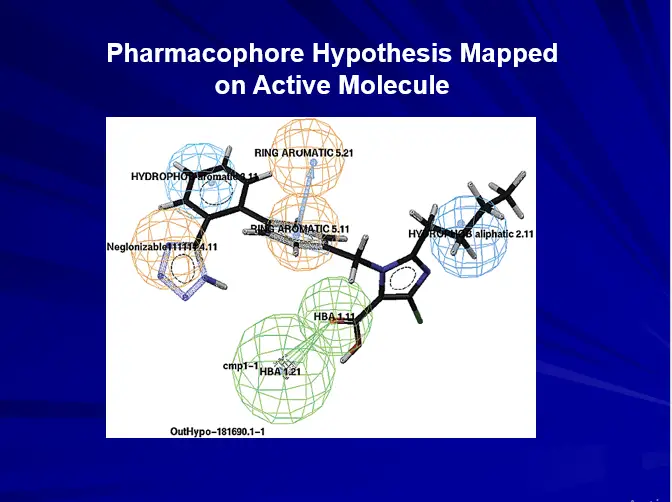
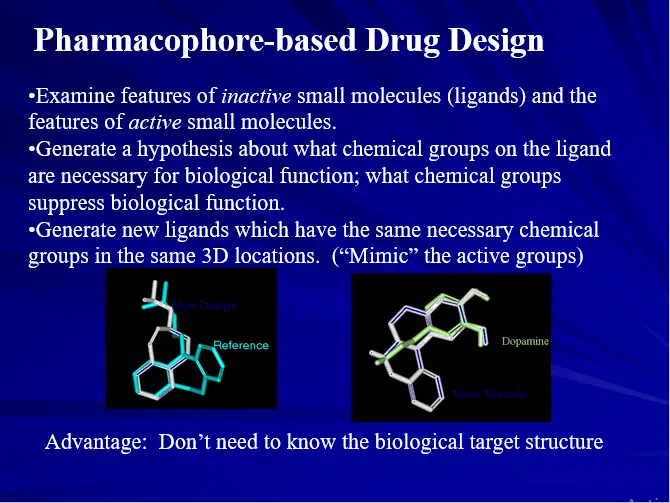
PHARMACOPHORE APPROCH
Pharmacophore:
The Spatial orientation of various functional groups or features in 3D necessary to show biological activity.
Types of Pharmacophore Models
Distance Geometry based Qualitative Common Feature Hypothesis.
Quantitative Predictive Pharmacophores from a training set with known biological activities.
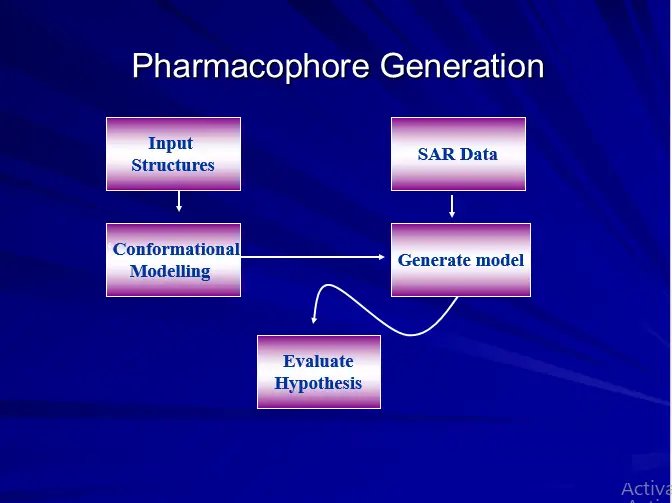
Five Steps
Training set selection.
Features selection
Conformation Generation
Common feature Alignments
Validation
Considerations/Assumptions
Training Set Molecules should be
– Diverse in structure
– Contain maximum structural information.
– Most potent within series.
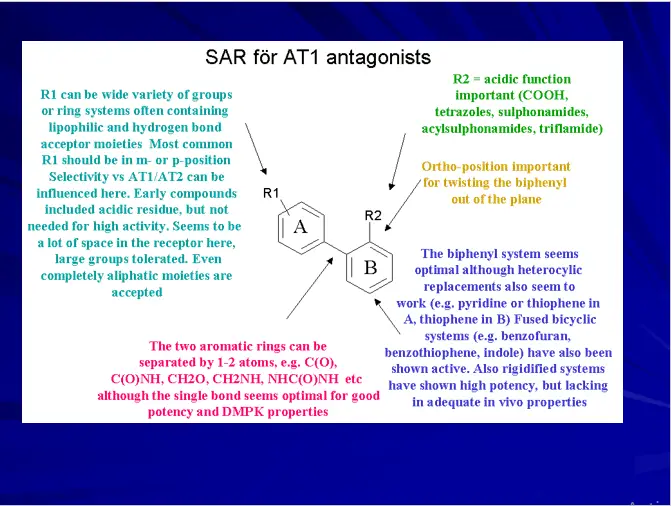
Features should be selected on the basis of SAR studies of training set
Each training set molecule should be represented by a set of low energy conformations. Conformations generation technique ensures broad coverage of conformational space.
Align the active conformations of the training set molecules to find the best overlay of the corresponding features.
Judge by statistical profile & visual inspection of model.
Pharmacophore Features
HB Acceptor & HB Donor
Hydrophobic
Hydrophobic aliphatic
Hydrophobic aromatic
Positive charge/Pos. Ionizable
Negative charge/Neg. Ionizable
Ring Aromatic
Each feature consists of four parts:
- Chemical function
- Location and orientation in 3D space
- Tolerance in location
- Weight
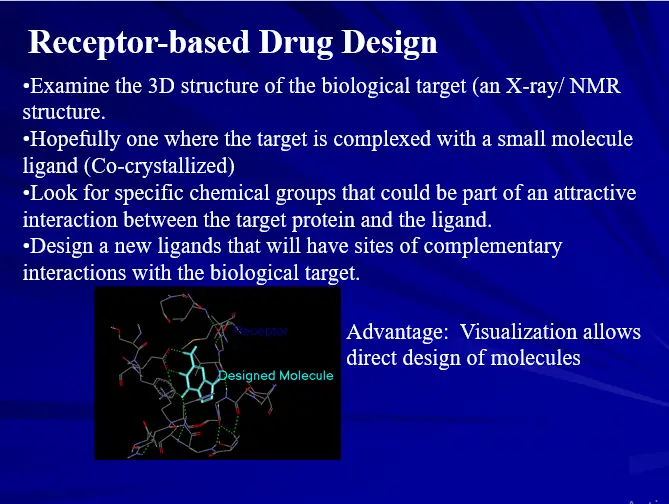
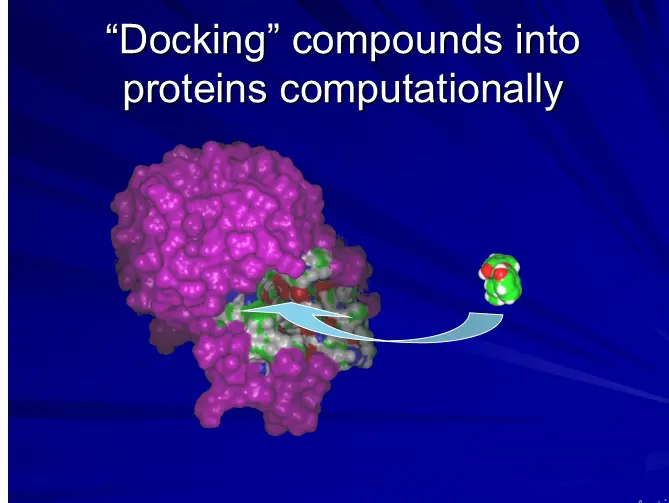
Docking Process
Put a compound in the approximate area where binding occurs
Docking algorithm encodes orientation of compound and conformations.
Optimize binding to protein
–Minimize energy
–Hydrogen bonding
–Hydrophobic interactions
Scoring
Rational Drug Design
Rational drug design refers to the development of medications based on the study of the structures and functions of target molecules. That is to say, the role of rational drug design is to use a methodological approach to coming up with a new drug, as opposed to blindly hoping some stroke of luck helps design a new drug, or instead of randomly testing hundreds of drug molecules in hopes that one of them binds to a receptor and exerts a therapeutic effect.
During rational drug design, researchers take three general steps to create a new drug:
Step 1. Identify a receptor or enzyme that is relevant to a disease they are going to design a drug for.
Step 2. Elucidate the structure and function of this receptor or enzyme.
Step 3. Use the information from step two in order to design a drug molecule that interacts with the receptor or enzyme in a therapeutically beneficial way.
This seems easy, but boy does it have to take a lot of important factors into account. For example, researchers must account for whether or not the drug will bind to a specific molecule or more than one. Binding to other molecules might bring about more adverse effects. How well the drug will bind to the target molecule needs to be clarified. If it can’t bind to the target well, it might be useless.
The shape and charge of the molecule must be examined in order to determine how this might affect the way it not only binds to the target, but is absorbed, distributed, metabolized, and excreted by the body.